Abstract
Background: Minimal residual disease (MRD) monitoring by PCR methods is a strong and standardized predictor of clinical outcome in mantle cell (MCL) and follicular (FL) lymphoma [Pott C., Blood 2010; Ladetto M, Blood 2013]. However, some technical limitations hamper its feasibility in daily clinical routine. First of all, current techniques allow the identification of a molecular marker only in 75-80% of MCL and 55-60% of FL patients. All the other "no marker" cases cannot be studied for MRD purposes. Next generation sequencing (NGS) tools might overcome these limitations. Among them, the recently developed targeted locus amplification (TLA) technology [de Vree P.J., Nat Biotechnol 2014] that selectively amplifies and sequences entire genes on the basis of the crosslinking of physically proximal DNA loci, was able to identify novel candidate oncogenes and gene fusions in acute lymphoblastic leukemia. [Kuiper R.P.; Vroegindeweij EM, 57th ASH Communication, Blood 2015].
The aim of this study was to test the TLA approach in MCL and FL diagnostic samples, with at least one translocation confirmed by FISH and a failure in molecular marker identification, with the aim to increase the rate of success in marker screening for MRD purposes. Moreover, the performances of the newly TLA molecular markers were compared to the standardized quantitative PCR (qPCR) approach for MRD monitoring.
Methods: Genomic DNA was extracted from bone marrow (BM) and peripheral blood (PB) samples of MCL and FL patients, enrolled in prospective clinical trials of the Fondazione Italiana Linfomi (FIL). All patients provided written informed consent for the research use of the biological samples. Libraries for TLA (Cergentis, Utrecht) were prepared using only one couple of antisense primers targeting the immunoglobulin heavy chain (IGH) enhancer locus, as previously described [Hottentot QP, Methods Mol Biol. 2017], and sequenced on MiSeq platform (Illumina, San Diego). MRD monitoring was carried out by qPCR designing allele specific oligonucleotides (ASO) and consensus probes based on the new breakpoint sequences obtained from TLA NGS. Finally, the efficiency of TLA novel markers to monitor MRD was compared to MRD data obtained from clonal IGH rearrangements detected and tracked by standardized ASO qPCR. [Van der Velden VHJ, Leukemia 2007].
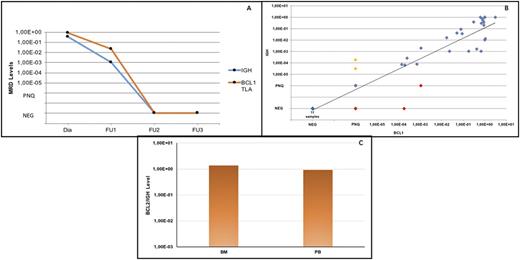
Results: TLA was firstly tested on 21 t(11;14)-positive, BCL-1/IGH major translocation cluster (MTC) negative MCL baseline samples (8 BM and 13 PB): in 86% of cases (18/21) a novel BCL-1/IGH breakpoint was identified by NGS. In these cases, TLA described new rearrangements between "no-MTC" BCL1 loci and JH1, JH4, JH5 or JH6 regions on chromosome 14. Therefore, additional 8 FL baseline BM samples, resulting t(14;18) FISH positive and BCL-2/IGH MBR/mcr/"minor rearrangements"-negative by PCR, were tested obtaining a 62% (5 out of 8 samples) success rate. Also in these cases, new and not yet described BCL2/IGH rearrangements were found. Additionally, to assess the specificity of the technology, the MBR-positive control sample (DOHH2 cell line) was tested and correctly sequenced, as well. Interestingly, among the new TLA positive FL cases, in one sample, the NGS technology highlighted the breakpoint on the DH region of chromosome 14, describing an unusual t(14;18) translocation. 5/29 cases (17%) in which TLA was not able to identify a new marker were characterized by a low tumor infiltration, mainly <5% by flow cytometry (median 4.4%, range 0.03-8%). Secondly, a validation of the NGS sequences was performed by ASO primers designed on the new TLA breakpoints and MRD was monitored by ASO qPCR. Overall, in 14 MCL cases where also a classic IGH-based marker was available, the MRD results were superimposable between the two markers (Fig1A), with good and overlapping performances in terms of overall correlation (r2=0,86; Fig 1B). In FL samples, ASO qPCR confirmed the high tumor burden of the analyzed tissues (Fig 1C). Finally, an extension of the analyzed samples series and MRD validation is currently ongoing among additional FL patients.
Conclusion: The TLA NGS technology allowed to identify a translocation-based novel molecular marker in FISH positive t(11;14) and t(14;18) MCL and FL baseline samples, where the classical Sanger sequencing failed a marker identification. These new breakpoints can reliably be used for the design of ASO qPCR primers for MRD detection in previously not traceable patients
Klous: Cergentis BV: Employment. Yilmaz: Cergentis BV: Employment. Vitolo: Gilead: Honoraria; Mundipharma: Honoraria; Roche: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Takeda: Honoraria. Luminari: PFIZER: Speakers Bureau; GILEAD: Speakers Bureau; TEVA: Membership on an entity's Board of Directors or advisory committees; CELGENE: Honoraria, Membership on an entity's Board of Directors or advisory committees; ROCHE: Membership on an entity's Board of Directors or advisory committees; TAKEDA: Membership on an entity's Board of Directors or advisory committees. Federico: takeda: Honoraria, Research Funding. Splinter: Cergentis BV: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal